GAGE is a method for identification of plant spe
发布日期:2026-01-20 18:15 浏览次数:
GAGE is a method for identification of plant species based on whole genome analysis and genome editing
 https://www.nature.com/articles/s42003-022-03894-9
Whole genomes of plants should be ideal databases for their species identification, but unfortunately there was no such method before this exploration. Here we report a plant species identification method based on the whole Genome Analysis and Genome Editing (GAGE). GAGE searches for target sequences from the whole genome of the subject plant and specifically detects them by employing a CRISPR/Cas12a system. Similar to how Mendel chose Pisum sativum (pea), we selected Crocus sativus (saffron) to establish GAGE, in which we constructed a library containing all candidate target sequences. Taking a target sequence in the ITS2 region as an example, we confirmed the feasibility, specificity, and sensitivity of GAGE. Consequently, we succeeded in not only using GAGE to identify Cr. sativus and its adulterants, but also executing GAGE in the plants from different classes including angiosperms, gymnosperms, ferns, and lycophytes. This sensitive and rapid method is the first plant species identification method based on the whole genome and provides new insights into the application of the whole genome in species identification.
植物全基因组本应是物种鉴定的理想数据库,然而此前尚无相关方法。本研究提出一种基于全基因组分析与基因组编辑(GAGE)的植物物种鉴定策略:从受试植物全基因组中筛选靶序列,并借助 CRISPR/Cas12a 系统实现特异性检测。如同孟德尔选用豌豆,我们以 Crocus sativus(藏红花)为模式建立 GAGE,构建了涵盖全部候选靶序列的文库。以 ITS2 区一段靶序列为例,验证了 GAGE 的可行性、特异性与灵敏度。结果不仅成功区分藏红花及其掺伪品,还将 GAGE 扩展应用于不同类群植物——被子植物、裸子植物、蕨类及石松类。该灵敏、快速的方法为首例基于全基因组的植物物种鉴定技术,为全基因组在物种鉴定领域的应用提供了新思路。
https://www.nature.com/articles/s42003-022-03894-9
Whole genomes of plants should be ideal databases for their species identification, but unfortunately there was no such method before this exploration. Here we report a plant species identification method based on the whole Genome Analysis and Genome Editing (GAGE). GAGE searches for target sequences from the whole genome of the subject plant and specifically detects them by employing a CRISPR/Cas12a system. Similar to how Mendel chose Pisum sativum (pea), we selected Crocus sativus (saffron) to establish GAGE, in which we constructed a library containing all candidate target sequences. Taking a target sequence in the ITS2 region as an example, we confirmed the feasibility, specificity, and sensitivity of GAGE. Consequently, we succeeded in not only using GAGE to identify Cr. sativus and its adulterants, but also executing GAGE in the plants from different classes including angiosperms, gymnosperms, ferns, and lycophytes. This sensitive and rapid method is the first plant species identification method based on the whole genome and provides new insights into the application of the whole genome in species identification.
植物全基因组本应是物种鉴定的理想数据库,然而此前尚无相关方法。本研究提出一种基于全基因组分析与基因组编辑(GAGE)的植物物种鉴定策略:从受试植物全基因组中筛选靶序列,并借助 CRISPR/Cas12a 系统实现特异性检测。如同孟德尔选用豌豆,我们以 Crocus sativus(藏红花)为模式建立 GAGE,构建了涵盖全部候选靶序列的文库。以 ITS2 区一段靶序列为例,验证了 GAGE 的可行性、特异性与灵敏度。结果不仅成功区分藏红花及其掺伪品,还将 GAGE 扩展应用于不同类群植物——被子植物、裸子植物、蕨类及石松类。该灵敏、快速的方法为首例基于全基因组的植物物种鉴定技术,为全基因组在物种鉴定领域的应用提供了新思路。
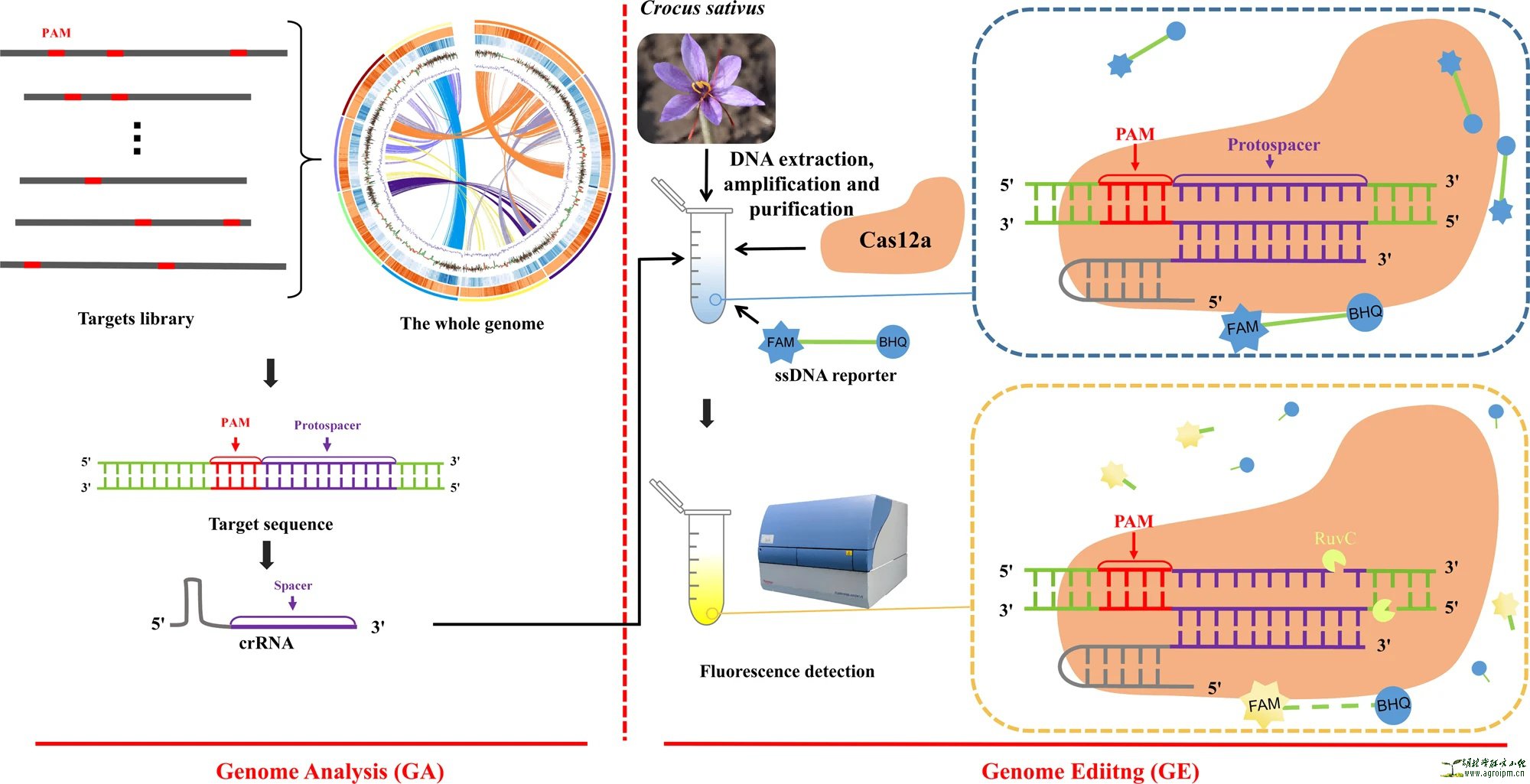 The fluorophore (6-Carboxyfluorescein, FAM) is linked to the quencher (Black Hole Quencher-1, BHQ) through ten dAs (ssDNA) and is initially quenched. After breaking away from BHQ, FAM absorbs light at its excitation wavelength and emits fluorescence. PAM: protospacer adjacent motif; The whole genome: The schematic genome draft was cited from the reference32; Targets library: The gather of target sequences; Target sequence: The sequence with a nearby PAM; crRNA: CRISPR RNA; ssDNA reporter: single-stranded DNA reporter (5’-FAM-AAAAAAAAAA-BHQ-3’, Poly_A_FQ).
The fluorophore (6-Carboxyfluorescein, FAM) is linked to the quencher (Black Hole Quencher-1, BHQ) through ten dAs (ssDNA) and is initially quenched. After breaking away from BHQ, FAM absorbs light at its excitation wavelength and emits fluorescence. PAM: protospacer adjacent motif; The whole genome: The schematic genome draft was cited from the reference32; Targets library: The gather of target sequences; Target sequence: The sequence with a nearby PAM; crRNA: CRISPR RNA; ssDNA reporter: single-stranded DNA reporter (5’-FAM-AAAAAAAAAA-BHQ-3’, Poly_A_FQ).

