Chromosome-level genome assembly provides insights into the
发布日期:2026-01-20 21:52 浏览次数:
Chromosome-level genome assembly provides insights into the genome evolution and functional importance of the phenylpropanoid–flavonoid pathway in Thymus mongolicus
https://doi.org/10.1186/s12864-024-10202-8
Background
Thymus mongolicus (family Lamiaceae) is a Thyme subshrub with strong aroma and remarkable environmental adaptability. Limited genomic information limits the use of this plant.
Results
Chromosome-level 605.2 Mb genome of T. mongolicus was generated, with 96.28% anchored to 12 pseudochromosomes. The repetitive sequences were dominant, accounting for 70.98%, and 32,593 protein-coding genes were predicted. Synteny analysis revealed that Lamiaceae species generally underwent two rounds of whole genome duplication; moreover, species-specific genome duplication was identified. A recent LTR retrotransposon burst and tandem duplication might play important roles in the formation of the Thymus genome. Using comparative genomic analysis, phylogenetic tree of seven Lamiaceae species was constructed, which revealed that Thyme plants evolved recently in the family. Under the phylogenetic framework, we performed functional enrichment analysis of the genes on nodes that contained the most gene duplication events (> 50% support) and of relevant significant expanded gene families. These genes were highly associated with environmental adaptation and biosynthesis of secondary metabolites. Combined transcriptome and metabolome analyses revealed that Peroxidases, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferases, and 4-coumarate-CoA ligases genes were the essential regulators of the phenylpropanoid–flavonoid pathway. Their catalytic products (e.g., apigenin, naringenin chalcone, and several apigenin-related compounds) might be responsible for the environmental tolerance and aromatic properties of T. mongolicus.
Conclusion
This study enhanced the understanding of the genomic evolution of T. mongolicus, enabling further exploration of its unique traits and applications, and contributed to the understanding of Lamiaceae genomics and evolutionary biology.
背景
百里香(Thymus mongolicus,唇形科)是一种具强烈香气且环境适应性突出的百里香属亚灌木。其基因组信息匮乏,限制了该植物的开发利用。
结果
构建了百里香染色体级 605.2 Mb 基因组,96.28% 的序列被锚定至 12 条假染色体。重复序列占主导地位,比例达 70.98%;共预测到 32,593 个蛋白编码基因。共线性分析表明,唇形科植物普遍经历两轮全基因组复制,并发现一次物种特异性基因组复制事件。近期 LTR 反转录转座子爆发及串联重复可能在百里香基因组形成中起关键作用。基于比较基因组学,构建了 7 种唇形科植物的系统发育树,显示百里香属在科内为近期分化。在该系统发育框架下,对支持度 > 50%、含最多基因复制事件的节点基因及显著扩增基因家族进行功能富集分析,发现这些基因与环境适应及次生代谢产物合成高度相关。转录组与代谢组联合分析揭示,过氧化物酶、羟基肉桂酰-CoA 莽草酸/奎宁酸羟基肉桂酰转移酶及 4-香豆酸-CoA 连接酶基因是苯丙烷-类黄酮途径的核心调控因子,其催化产物(如芹菜素、柚皮素查尔酮及多种芹菜素相关化合物)可能赋予蒙古百里香环境耐受性与芳香特性。
结论
本研究深化了对百里香基因组演化的认识,为其独特性状挖掘与应用奠定基础,同时为唇形科基因组学与进化生物学研究提供新见解。
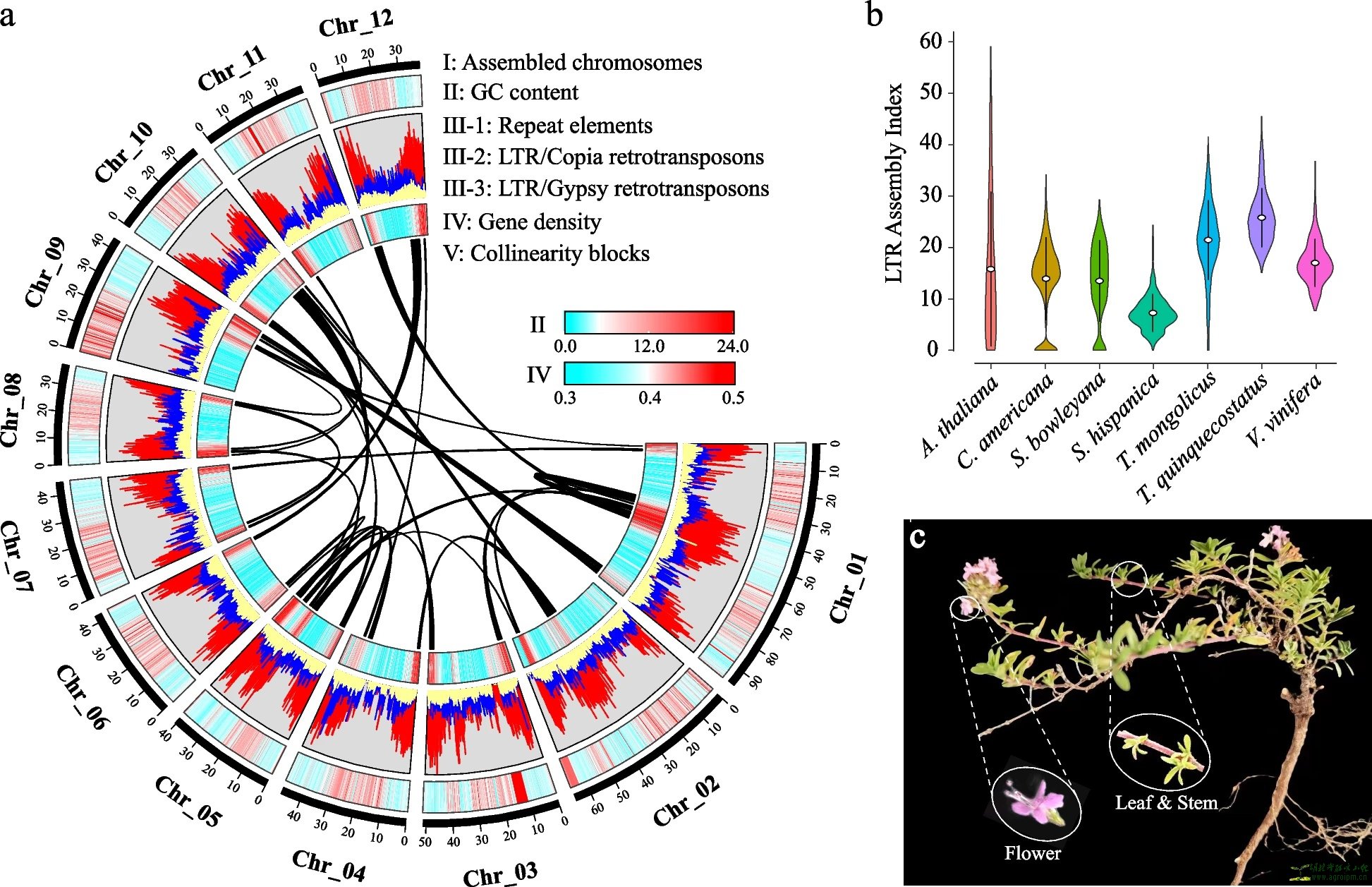
The features of Thymus mongolicus genome. a The landscape of T. mongolicus genome. The circus plot from the outer to the inner circle represents chromosome-scale pseudochromosomes (Chr01–Chr12) (I), GC content (II), repeat element (III-1), Ty1/copia retrotransposons (III-2), Ty3/gypsy retrotransposons (III-3), gene density (IV), and each linking line in the center of the circus plot indicates the collinearity blocks panning more than 40 genes in the genome (V). b LTR assembly index (LAI) analysis for the referenced genomes. c T. mongolicus plant

