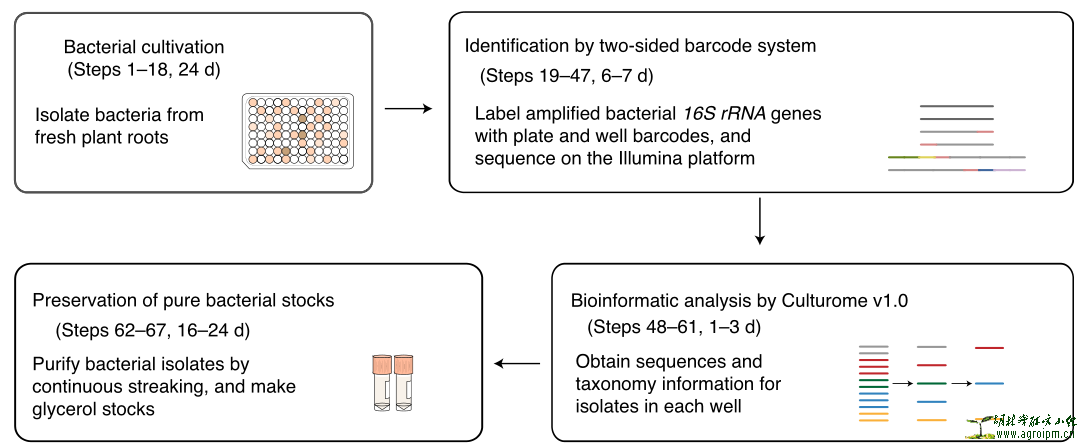
High-throughput cultivation and identification of bacteria f
发布日期:2022-06-10 15:01
来源:Nature Protocols
作者:Yang Bai
浏览次数:
Abstract
Cultivating native bacteria from roots of plants grown in a given environment is essential for dissecting the functions of the root microbiota for plant growth and health with strain-specific resolution. In this study, we established a straightforward protocol for high-throughput bacterial isolation from fresh root samples using limiting dilution to ensure that most cultured bacteria originated from only one microorganism. This is followed by strain characterization using a two-sided barcode polymerase chain reaction system to identify pure and heterogeneous bacterial cultures. Our approach overcomes multiple difficulties of traditional bacterial isolation and identification methods, such as obtaining bacteria with diverse growth rates while greatly increasing throughput. To facilitate data processing, we developed an easy-to-use bioinformatic pipeline called ‘Culturome’ (https://github.com/YongxinLiu/Culturome) and a graphical user interface web server (http://bailab.genetics.ac.cn/culturome/). This protocol allows any research group (two or three lab members without expertise in bioinformatics) to systematically cultivate root-associated bacteria within 8–9 weeks.
高通量分离与鉴定筛选基本流程
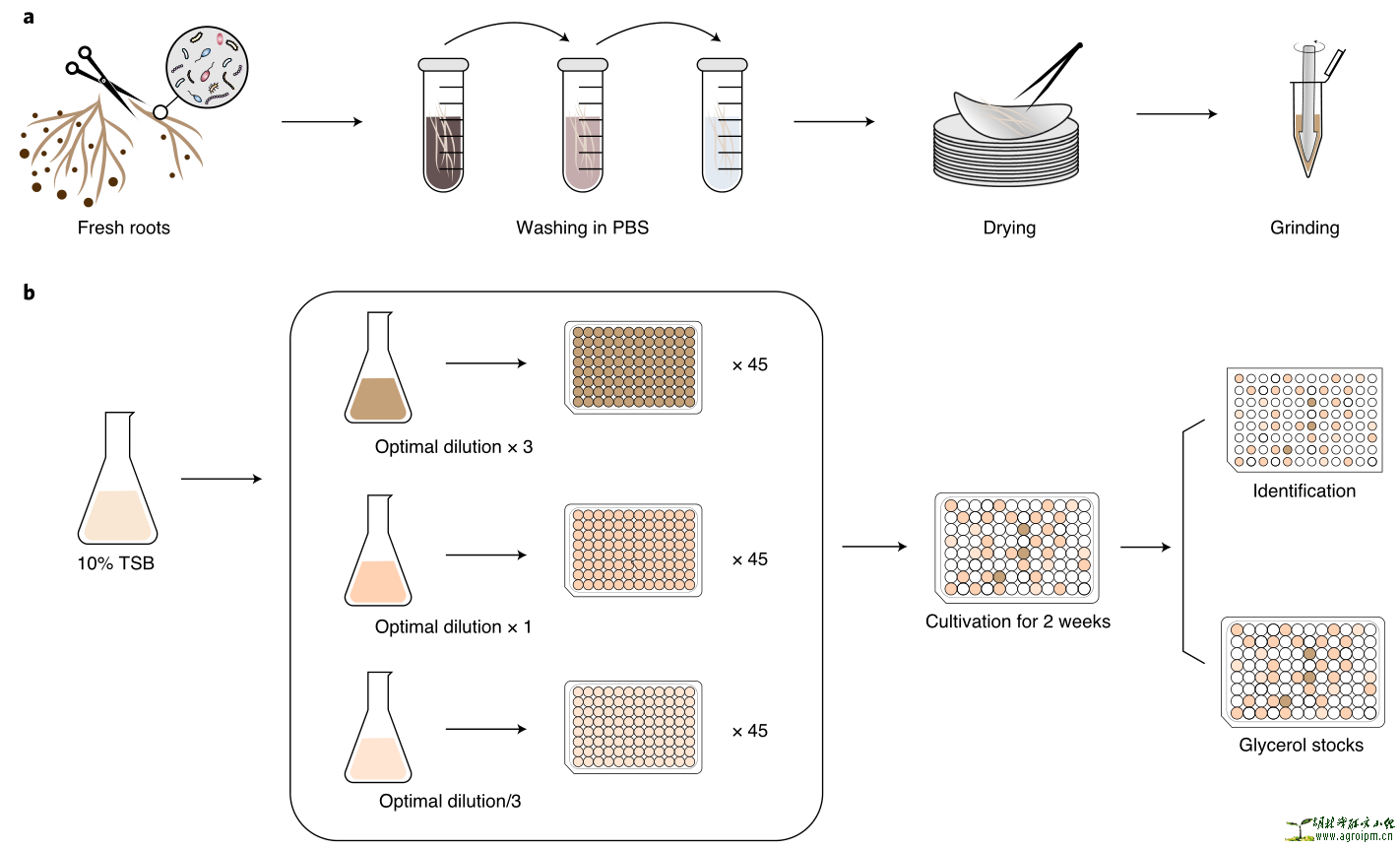
 高通量筛选方法
高通量筛选方法

“Click” reactions in polysacchari
2015-11-13
Xiangtao Meng a , b , Kevin J. Edgar a , b , , a Macromolecules and Interfaces Institute, Virginia Tech, Blacksburg, VA 24061, United States b Department of Sustainable Biomaterials...

中国丝状真菌次级代谢分子调
2019-04-18
遗传, 2018, 40(10): 874-887 doi: 10.16288/j.yczz.18-169 综述 中国丝状真菌次级代谢分子调控研究进展 潘园园,1, 刘钢,1,2 1. 中国科学院微生物研究所,真...

Self-Fueled Biomimetic Liquid Metal M
2015-03-27
Self-Fueled Biomimetic Liquid Metal Mollusk Authors Jie Zhang, Youyou Yao, Lei Sheng, Jing Liu Abstract A liquid metal motor that can eat aluminum food and then move spontaneously a...
A New Golden Age of Natural Products
2016-07-23
Ben Shen DOI: http://dx.doi.org/10.1016/j.cell.2015.11.031 The 2015 Nobel Prize in Physiology or Medicine has been awarded to William C. Campbell, Satoshi Omura, and Youyou Tu for t...






